DNA Alignment Help | Editing example: How to cure insertion artefacts
This example demonstrates how to use the manual editing mode.
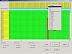
Please look briefly at the webpage for Fig. 1 and then return to this page. In this example, the taxon Nuu10 has an insertion artefact between position 16362 and position 16368. This was an occasional error by early versions of DNA_Alignment (which should not occur today in current versions, when the sequences in the data set are closely related). The desired optimal alignment is shown in the last figure (see Fig.7 in full size).
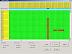
To correct the insertion, move the mouse onto position 16362 of Nuu10 and press the right mouse button. A pop-up window with commands will appear (see Fig.2 in full size). Select "Delete" to delete the "-". (Alternatively, press the delete key on the keyboard). For taxon Nuu10, this shifts the contents of positions 16363ff left by 1 cell, deleting the cell content "-" at position 16362. The result is shown in Fig.3 (see Fig.3 in full size).
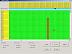
Next, insert a "-" to the right of position 16367, for taxon Nuu10 (see Fig.4 in full size). Use the pop-up menu or alternatively the keyboard key "insert". This will insert a "-" and shift the cell contents "TGGAT..." of Nuu10 to the right by 1 cell. The result is shown in Fig.5 (see Fig.5 in full size).
Finally, inspection shows that the entire column "-" between positions 16367 and 16368 is now superfluous and optionally can be deleted to give a neater table. To do this, move the mouse onto one of the yellow top rows and press the right mouse button (see Fig.6 in full size). Select "delete". The result is shown in Fig.7 (see Fig.7 in full size).
For users who prefer the keyboard to the mouse, see keyboard functions.