Network | Release notes
Current Release: Network 10.2.0.0, released 2020 December 30
1. Due to user wishes, the limit of characters (e.g. nucleotide positions) is increased to 35000 to allow users to more easily create SARS-CoV-2 rdf files with their own software or scripts. But please note that we do not have the human resources to assist users to program, test, or debug their software or scripts.
2. New Beta-Version of VCF2RDFconverter.exe to convert VCF files to binary and/or multistate RDF files. This converter is "work in progress", and we include it because it may be useful. But you will need to manually check your VCF file and also the resulting RDF file for the following known bugs: (1) Deletions will not be converted correctly and the character is usually omitted entirely from the sequence in the RDF file even though the character is present in the RDF header. (2) If the VCF file contains two or more lines with the same genomic coordinate but different REF/ALT values, then the character name in the RDF will need to be checked by hand and possibly edited. We welcome bug reports if you include a very small example file to help us reproduce your bug.
3. User guide update with some advice how to obtain SARS-CoV-2 mutations from next generation sequencing (NGS) data. Importantly: Primer regions in amplicons (e.g. ARTIC primers) are susceptible to primer errors. With appropriate software and software settings, these primer artefacts can be avoided. We explain how. In long range PCR amplicons (e.g. Eden primers), even non-primer regions may show artefacts, so we would suggest that technical replicates of the long range amplicons are done and sequenced independently.
Known issues in Network 10.2.0.0.:
1. VCF2RDFconverter.exe has bugs and the output RDF files need to be manually checked/edited: see above.
2. Network does not show any warning message when failing to overwrite write-protected files, or when failing to write files into a write-protected folder. In the data editor, for these cases the wrong message "File saved. [...]" is displayed. Work-around suggestions: After saving, check the time and date of the file which you have saved. Before saving, if you are uncertain whether a folder or a file is write-protected, right-click the folder or file, select "properties", and see whether write protection is checked.
3. Display problems on small displays or low-resolution (laptops): Buttons at bottom not visible / below the Windows task bar. Work-arounds: use free Network 4.6.1.6 or configure the Windows task bar to hide itself automatically or increase the display resolution (if possible) or reduce the size of text and apps in Windows Display / Scale and layout.
Improvements from Network 10.0.0.0 / Network 5.0.1.1.CS to Network 10.1.0.0 (released 2020 April 18)
1. This version replaced Network 10.0.0.0 and Network 5.1.1.1.CS
2. Re-instated Postprocessing option MP-Postprocessing (by Polzin and Daneshmand) that could not be included in the previous version.
3. Fixed the "Weighted length of Steiner trees" value (in Network Draw / Statistics) or "Cost" value (in the MP *.sto file). Compared to the previous version, this fix only changes the reported and displayed value but not the MP computation or the Network results.
Improvements from Network 5.0.1.1 to Network 10.0.0.0 / Network 5.0.1.1.CS (released 2020 January 18)
1. Fixed Network 5 to run on Windows 10 without the previous nagging warning messages.
Improvements from Network 5.0.1.0 to Network 5.0.1.1 (released 2019 January 13):
1. Fixed the new feature in Draw Network that led to crash when loading some files, esp. when loading large networks: In the "Display parameters" control panel, new checkbox "Display mutated position lines". When enabled, tick-marks are drawn on the links to symbolize the markers (characters) on each link. This enables classical network graphs to be generated without displaying the name of the mutated position.
Improvements from Network 5.0.0.3 to Network 5.0.1.0 (released 2018 December 17):
1. New feature: VCF2RDFconverter. This is a small stand-alone app to convert VCF files (variant call format files from next-generation sequencing data analysis) to Network's binary RDF format. This new-format binary RDF file will only work with Network 5.0.1.0, not with legacy Network 4.6.1.6. Before converting a VCF-file to RDF format, please make sure that the VCF file contains only non-recombining data and that the number of markers is less than the Network limits.
Non-recombining data that are appropriate for phylogenetic network analysis include: (1) bacterial or virus genomic markers, (2) mitochondrial DNA markers in otherwise diploid organisms (maternal lineage), (3) y-chromosomal markers in otherwise diploid organisms (paternal lineage), (4) somatic mutation analysis within a cancer patient between germline, tumor and metastasis sequence data (see ExampleMeissner2017 precision medicine cancer evolution phylogeny files in Network download, Meissner et al 2017, PMID: 28679691).
To improve the quality of your analysis, it is recommended to use only the highly confident markers. Ideally, all variants or somatic mutations should be validated by manual inspection of the alignments in the BAM files, e.g. using IGV. As Network requires short names (for more clarity in the network graph display), VCF2RDFconverter renames genomic positions with short numbers, beginning at 1. The translation table between these short numbers and the original genomic positions can be looked up in lines 2 and 3 of the RDF file (which is in text format).
2. New Feature in Draw Network: In the "Display parameters" control panel, new checkbox "Display mutated position lines". When enabled, tick-marks are drawn on the links to symbolize the markers (characters) on each link. This enables classical network graphs to be generated without displaying the name of the mutated position.
Improvements from Network 5.0.0.1 to Network 5.0.0.3 (released 2017 December 21):
1. Minor user interface updates.
2. ExampleMeissner2017 precision medicine cancer evolution phylogeny files in Network download. Meissner et al 2017 Metastatic triple-negative breast cancer patient with TP53 tumor mutation experienced 11 months progression-free survival on bortezomib monotherapy without adverse events after ending standard treatments with grade 3 adverse events, PMID: 28679691.
Improvements from Network 5.0.0.0 to Network 5.0.0.1 (released 2016 December 23):
1. Time estimates: Fixed layout of user interface.
2. Tools: Fixed layout of mismatch distribution tool user interface.
Improvements from Network 4.6.1.3 to Network 5.0.0.0 (released 2015 December 24):
1. Major user interface updates to Data Entry, Network Calculations, and Draw Network. These updates solve display formatting problems.
2. Major software port from an old free software compiler to a new non-free software compiler. The network calculation results are all identical to the old version's results. Compiler-related memory problems are much improved using this new commercial compiler, i.e. larger data sizes should be possible.
Network 4 (fall-back releases): Network 4.6.1.4./4.6.1.5., released 2015 December 24 / 2016 December 23
1. Minor user interface updates.
Known issues in Network 4.6.1.4./4.6.1.5.:
1. Network does not show any warning message when failing to overwrite write-protected files, or when failing to write files into a write-protected folder. In the data editor, for these cases the wrong message "File saved. [...]" is displayed. Work-around suggestions: After saving, check the time and date of the file which you have saved. Before saving, if you are uncertain whether a folder or a file is write-protected, right-click the folder or file, select "properties", and see whether write protection is checked.
2. Time estimates: A node colouring bug remains in this release of Time estimates. The nodes are not displayed as defined in Network Publisher or Network Draw. When a pie-slice node is selected, only part of this node is highlighted.
Improvements from Network 4.6.1.2 to Network 4.6.1.3. (released 2014 December 24):
1. Minor user interface updates.
Improvements from Network 4.6.1.1 to Network 4.6.1.2. (released 2014 January 1):
1. Minor user interface updates.
Improvements from Network 4.6.1.0 to Network 4.6.1.1. (released 2012 December 20):
1. New Feature: In Network Draw, new option to deactivate median vector names.
Improvements from Network 4.6.0.0 to Network 4.6.1.0. (released 2011 December 30):
1. New Feature: "MJ square" option in the Median-Joining network calculation ("Parameters" menu) can identify potential MP-links which were not previously identified, at the cost of longer run times. By default, this new option is switched off. The new option is likely to be of interest primarily in small and clear data sets, because large data sets will require star-contraction preprocessing and/or the frequency>1 criterion to make them more legible.
2. Fix: Draw Network / Shortest Trees box: After MP-cleanup and loading the *.sto file, the radio button "Original Network" now correctly displays the original network.
Improvements from Network 4.5.1.6 to Network 4.6.0.0. (released 2010 December 31):
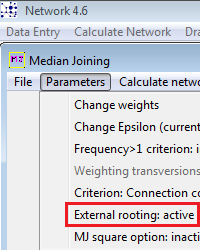
1. New Feature: External rooting using an outgroup sequence named ROOT to find the root proxy node (nearest network node to the outgroup): Activate this calculation option in Calculate Network / Median Joining network calculation / Parameters menu.
2. New Feature: Network rdf editor: New button "Change multiple weights" to assign the same weight to a range of nucleotides.
3. Median Joining network calculation of star-contracted amino acid data: Fixed issue with sco-files which were generated from ami-files.
4. Some warning/error messages were added and misleading messages were corrected.
5. Expired Network version: New button "Download newest Network version" in the "About/Contact" menu.
Improvements from Network 4.5.1.0 to Network 4.5.1.6. (released 2009 December 31):
1. Larger file and data sizes: Numerous out-of-memory bottlenecks were addressed in Median-Joining network calculation, in Reduced-Median network calculation, and in Draw. For median-joining calculations, convergence runs with different epsilon-values are possible which previously hit the limits. Caveat: Limits were hit due to system heap (not RAM), therefore the software code was re-written in many places. You may still encounter system heap memory limits with some data sets.
2. RFLP (*.tor) file format: Limit extended to 5000 polymorphisms.
3. Corrupted or wrong files: Auto-binarisation of multistate rdf files when loading into the RDF editor in binary mode, or into the Reduced Median network calculation. If it is not possible to auto-binarise the RDF file, there is a warning message. For corrupted files or unsupported formats, the error messages have been improved.
4. RDF/STR editor: Long taxon names or long locus names are displayed in full length when the mouse-tip is moved over the cell. Warning and error messages have been improved.
5. Star contraction: New format (beta): Amino acid data (*.ami). Note that star contraction on the *.ami format has only been tested on small test data sets - please QC your results accordingly.
6. Reduced Median network calculation: New checkbox for complete calculation (default: ON) or stopping after saving the rmf file. If run times are too long, or if memory problems are encountered during the complete RM calculation run, the rmf file can be used for a subsequent Median Joining network calculation (which runs faster and can handle larger data sets than RM). See Network calculation / Reduced Median / Parameters menu / reduction threshold.
7. Draw: Larger data sets (see 1.) and improved link selection in complex networks (previously very difficult).
8. Time estimates: Improved handling of large or complex networks: More control over the displayed graphics is available. A minor problem has been fixed when only two median vectors are selected.
Improvements from Network 4.5.0.2 to Network 4.5.1.0. (released 2008 December 27):
1. New Feature: Network rdf editor, dna data mode: New button to binarise dna data, so that the RM network calculation may be run for a "second opinion".
2. New Feature: Network rdf/STR editor, extended rdf and ych formats: Additional information can be entered for taxa/sequences (and used in Network Publisher for network pie-coloring). Instead of "Ethnic Group, Region, Haplogroup", the first 3 fields are now corrected to "Phenotype, Geography, Lineage". Three additional fields "Group 1, Group 2, Group 3" are available, which can be user-renamed in Network Publisher.
3. New Feature: Network Draw: Double-click on node opens new node-info pop-up window with extended node information including long taxon names and additional information (phenotype, geography, etc).
4. Fix: Network calculation (RM, MJ, or star contraction) importing phy, nex, tor, or ych format: warning if rdf file with same name exists, so that no data is overwritten without warning.
5. Fix: When opening a file, in extremely rare cases this file was deleted. We think that this problem only occured when Network was run on PCs with specific unfortunate combinations of older Windows service packs and older hotfixes. However, to prevent data loss, Network now opens a copy (file Interim_Sto.txt or Interim.txt) instead of the original file. The interim files are intentionally not deleted after Network is closed.
6. Fix: Network rdf/STR editor: Undo now undoes the function "Set default character state".
7. Fix: Network rdf/STR editor and Network calculations: Networks were correct, but pie charts incomplete: Did not import "additional information" of taxon if the preceding "additional information" fields were empty.
8. Fix: Network rdf/STR editor: In "additional information" mode (after "switch windows"), the delete-key will now delete entries, and under Windows 2000 copy/paste (CTRL+C/CTRL+V) now works.
9. Fix: Network rdf/STR editor: When hitting the exit button without having saved changes, the correct warning message is now displayed.
10. Fix: Network calculations, file with problems: After 5 file problem messages, the import is aborted with the warning message "Further errors in the file will not be displayed".
Improvements from Network 4.5.0.1 to Network 4.5.0.2 (released 2008 July 23):
1. Fix: window auto-fit for "wide" displays (used increasingly in new laptops).
Improvements from Network 4.5.0.0 to Network 4.5.0.1 (released 2008 June 24):
1. Fix, Star Contraction calculation: Long haplogroup names exported correctly.
2. Fix, Draw Subprogram: Load dia file and save correctly as fdi.
3. Fix, Network Data Editor: Fixed some screen resolution mode dependent problems in the Network rdf editor / str editor: cell-highlighting when paging down or up, alignment of the table cells with the cells in the name column, frequency column, locus header row, and weights footer row.
4. Fix, tor file into Star Contraction Calculation, Reduced Median Calculation, Median-Joining Calculation: Corrected tor file import for the special case of a data set of sequences, in which the same nucleotide position (with respect to the reference sequence) has been cut by different enzymes.
Improvements from Network 4.2.0.1 to Network 4.5.0.0 (released 2007 December 31):
1. Fixed occasional program crash problems in MP option (Polzin et al). MP is recommended after network calculations (see box).
2. New file format versions (rdf, ych, sco, out, sto, fdi). Network 4.5.0.0 can read the old files produced by earlier Network versions.
WARNING: Network 4.5.0.0 will SAVE (OVERWRITING OLD FILES with same name) in the NEW FORMAT, so BACK UP your OLD Network files! (In general it is good practise to make safety copies of files regularly)
3. Longer names: Names of characters / features / nucleotide-positions / RFLP-loci up to length 8, names of STR-loci up to length 6, names of individuals / sequences / groups / taxa / STR-profiles up to length 15.
WARNINGS: a) Auto-layout in Network Draw is different for some data, see note below. b) Network Draw only displays the first 6 characters of a name, because longer names tend to obscure the network graphics.
4. Weights for Y-STR/microsatellite/autosomal-STR/mixed data: Entry of weights, and saving to file (in new *.ych format) is now possible in the Network STR-editor (Data Entry menu, Manual or Import rdf file, Y-STR data).
5. Further information ("attributes") for each sequence/taxon/STR-profile: In Network 4.5.0.0, you can enter names of Haplogroups, Ethnic Groups, or Geographic Regions for each taxon/sequence/STR-profile in the Network data editor (Data Entry menu, Import rdf file, after import click "Switch input window" button). The attributes are saved in the new rdf and ych files, and can be loaded back into the Network editor for later reference. In the Network Publisher add-on software (this costs a fee), the attributes can be used for colouring network pies and nodes.
6. Fixed problem loading *.dia files into Network Draw. This problem occurred when Network4.2 was run on Windows versions with recent "Windows hotfixes" (Windows auto-updates).
7. For STR data with very large number of profiles: Up to 676 polymorphisms of each STR locus are now allowed (previously 26).
Note: Potential graphical layout changes due to new naming:
For users who notice differences in the automatic graphical layout of some networks (esp complex networks): The graphical layout of some Network 4.5.0.0. results is different compared to the graphical layout of Network 4.2.0.1 results. This is no error: The graphical layout differences are due to different naming in Network 4.5.0.0 (longer character names in STR calculation results, some lower case spelling such as "sco" instead of "SCO", etc), which influences the "preferred relatively empty regions" into which the network links are automatically drawn.







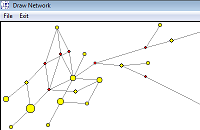
 With MP postprocessing (Polzin et al) after network calculation (*.sto file):
With MP postprocessing (Polzin et al) after network calculation (*.sto file):