DNA Alignment Help | Changing/selecting the reference sequence
Default reference sequence
By default, DNA Alignment uses the file "ReferenceSequence.txt" in its start-up folder as the reference sequence. In the DNA Alignment download, this file is the same as the file RefSeqHumanmtDNAcontrolregion.txt (human mtDNA control region HV1 and HV2). The file corresponds to FASTA-format, except for the optional first line. The optional first line specifies the nucleotide numbering of linear
DNA (chromosomes) and of circular DNA (e.g. mtDNA, plasmids).
Example line (from "ReferenceSequence.txt"):
<15996-16569;1-576
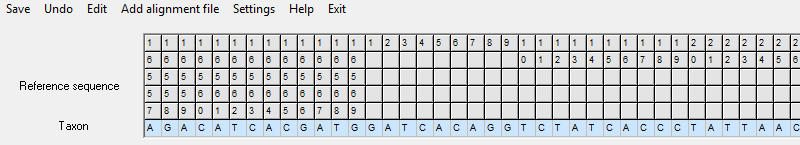
The reference sequence in the file is defined for nucleotide positions 15996-16569, followed by nucleotide positions 1-576. These nucleotide positions are displayed by the software, and the reference nucleotides are displayed under the positions:
 Changing the reference sequence
Changing the reference sequence
There are two ways to change the reference sequence:
-
If you need a particular reference sequence for most or all of your alignments: delete the original ReferenceSequence.txt file and rename a copy of your reference sequence file (in the DNA Alignment software folder) to "ReferenceSequence.txt".
-
If your reference sequences often change: manually select the reference sequence file before you load and auto-align the FASTA file of individual sequences. The reference sequence file may have any name provided that it has a *.txt extension. The manually selected reference sequence remains active until you select another reference sequence or until you exit the software. To align a file of sequences to the first sequence in this file: Manually select this file as the reference sequence, then load and align this same FASTA file.